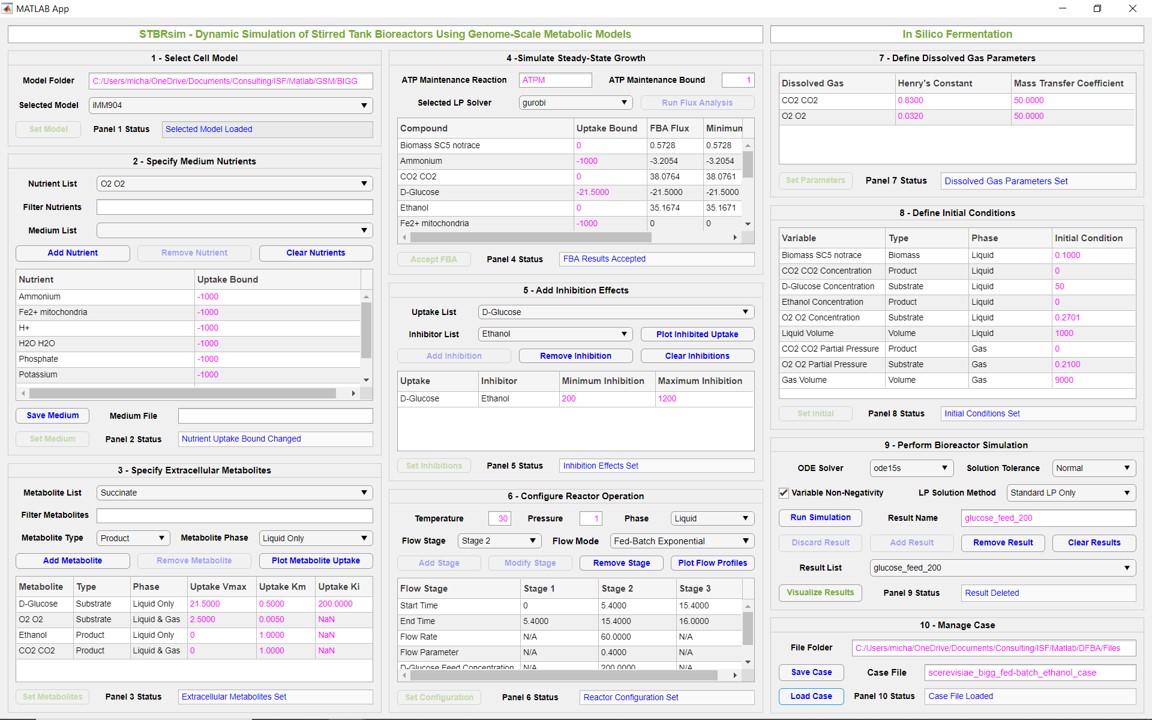
Snapshot of STBRsim for analyzing fed-batch feeding policies for ethanol production in wild-type Saccharomyces cerevisiae.
STBRsim version 2.0 was released by In Silico Fermentation in February 2025. Please continue reading this page to learn more about the application and to download this free software.
An emerging application of genome-scale metabolic reconstructions is the development of rigorous dynamic simulations of stirred tank bioreactors (STBRs). By exploiting dynamic flux balance analysis (DFBA) methods which integrate numerical solution of the intracellular flux linear program (LP) and the extracellular ordinary differential equations (ODEs), the same metabolic model can be used as a common platform for medium design, metabolic engineering and bioreactor simulation. While the research literature contains numerous DFBA computational methods, effective utilization of these approaches requires significant experience with high-level programming languages such as MATLAB. As a result, available modeling tools are largely inaccessible to non-expert researchers and practitioners who wish to perform bioreactor simulations with minimal coding effort.
STBRsim is a MATLAB GUI application that streamlines the development of dynamic metabolic models for bioreactor simulation. Given a genome-scale metabolic reconstruction of the target strain, the App generates time-resolved predictions of biomass, substrate and product concentrations, liquid/gas phase volumes, and intracellular-extracellular transport fluxes for user-specified inhibition effects, liquid/gas feeding strategies and reactor conditions. The workflow allows the definition of the growth medium (compatible with MediumFBA), growth-limiting substrates and secreted products along with their uptake kinetics, growth inhibition effects involving extracellular substrates and products, bioreactor operating conditions such as multiple stages for different liquid/gas feeding policies, dissolution and mass transfer parameters for dissolved gases, and initial conditions for the extracellular variables. Dynamic simulations can be performed for different combinations of the user-defined model parameters and stored for plotting and comparison using an included visualization App.
STBRsim has the following requirements:
- Version 2021a or newer version of MATLAB.
- Installed COBRA toolbox for MATLAB.
- A linear program (LP) solver compatible with COBRA. A list of compatible LP solvers is available here.
- A genome-scale metabolic reconstruction compatible with COBRA. STBRsim has been tested with selected reconstructions from the BIGG repository and the Virtual Metabolic Human resource (i.e., AGORA1 models).
STBRsim can be freely downloaded using the link below. The software is distributed under The GNU General Public License v3.0. Based on user feedback, Version 2.0 of STBRsim offers the following improvements over Version 1.1:
- Incorporation of growth inhibition effects where any substrate uptake rate can be inhibited by any extracellular substrate or product according to a piecewise linear inhibition function.
- Addition of plotting capabilities for substrate uptake rates to visualize both uninhibited and inhibited uptake rate functions.
- Addition of plotting capabilities for time-dependent liquid and gas flows to visualize the feed and outlet flow rate functions, the liquid and gas volumes, and the total amount of each substrate fed at any time point.
- Modification of bioreactor simulation options to provide additional flexibility with LP and ODE solution methods.
- General improvements in the App workflow and removal of a few assorted coding bugs.

STBRsim is a MATLAB application that performs dynamic bioreactor simulations based on genome-scale metabolic reconstructions.
The Zip file should be processed as follows:
- Download and unzip the file using Zip archive software.
- The unzipped file will contain the following files: (1) the main application, STBRsim_ver_2_0.mlappinstall; and (2) saved App states comparing alternative batch-to-continuous switching times for anaerobic growth of Escherichia coli (ecoli_agora_anaerobic_startup_case), alternative oxygen mass-transfer coefficients for glucose-to-acetate diauxic growth of E. coli (ecoli_bigg_aerobic_diauxic_case), and alternative glucose feed concentrations for fed-batch ethanol production from Saccharomyces cerevisiae (scerevisiae_bigg_fed-batch_ethanol_case).
- Save the files in the desired working directory.
- To install the STBRsim application, go to the Apps tab on the main Matlab page and click “Install App”. Select the saved STBRsim_ver_2_0.mlappinstall. The STBRsim application will appear as a new icon in the Apps tab.
- Launch the application by clicking on the icon.
Please contact me by email if you have any questions or feedback about STBRsim.