In addition to representing a powerful research paradigm, genome-scale metabolic modeling offers numerous opportunities as a platform for educational tool development. Models can be inconspicuously embedded within software tools to allow students access to the full capabilities of metabolic modeling without emphasis on the associated metabolic network properties, mathematical underpinnings and implementation details. Consequently, students can focus on learning microbial and biosystems engineering concepts that are the usual emphasis of their academic studies.
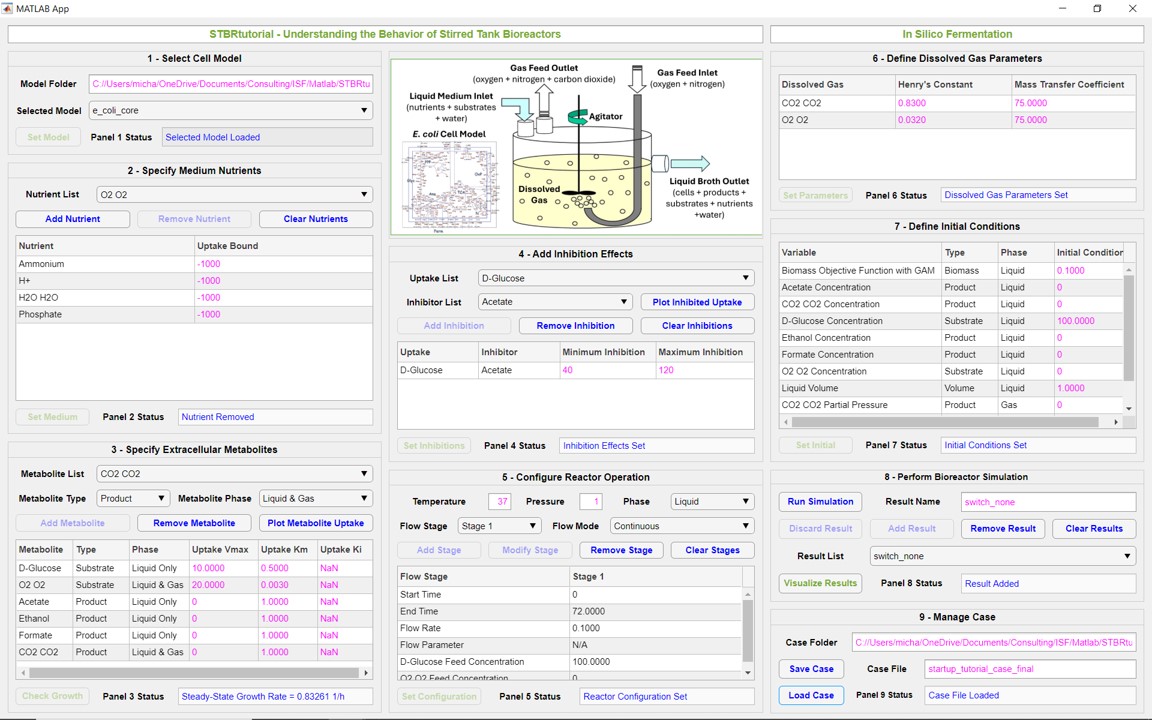
In Silico Fermentation is developing a suite of free MATLAB GUI applications that provide students with detailed tutorials that address important concepts in microbial systems engineering. These tutorials build upon and strategically simply our research Apps developed for industrial practitioners and academic researchers. The underlying philosophy is to minimize the metabolic modeling component so students can focus on higher level problems, such as medium design or bioreactor dynamics. The following tutorial Apps have been released or are under development.
- STBRtutorial – Understanding the Dynamic Behavior of Stirred Tank Bioreactors (first release January 2025)
- FBAtutorial – Understanding the Nutritional Requirements and Metabolic Behavior of Microbial Strains (first release anticipated Winter 2025)
These applications have been designed with seamless integration of metabolic modeling such that educators can provide their students will software training tools that address complex microbial systems engineering principles that are difficult to adequately cover in a standard lecture environment.