Genome-scale metabolic reconstructions have emerged as powerful computational platforms for analyzing and engineering microbial metabolism. Researchers typically develop their own workflows by writing customized code around general-purpose functions available within software tools such as Constraint-Based Reconstruction and Analysis Toolbox (COBRA). The development of effective and robust workflows can be challenging due to incompatibility between component functions, difficulty in integrating user-designed codes, and the substantial overhead associated with code development and testing. As a result, the power of metabolic modeling has been largely limited to domain experts.
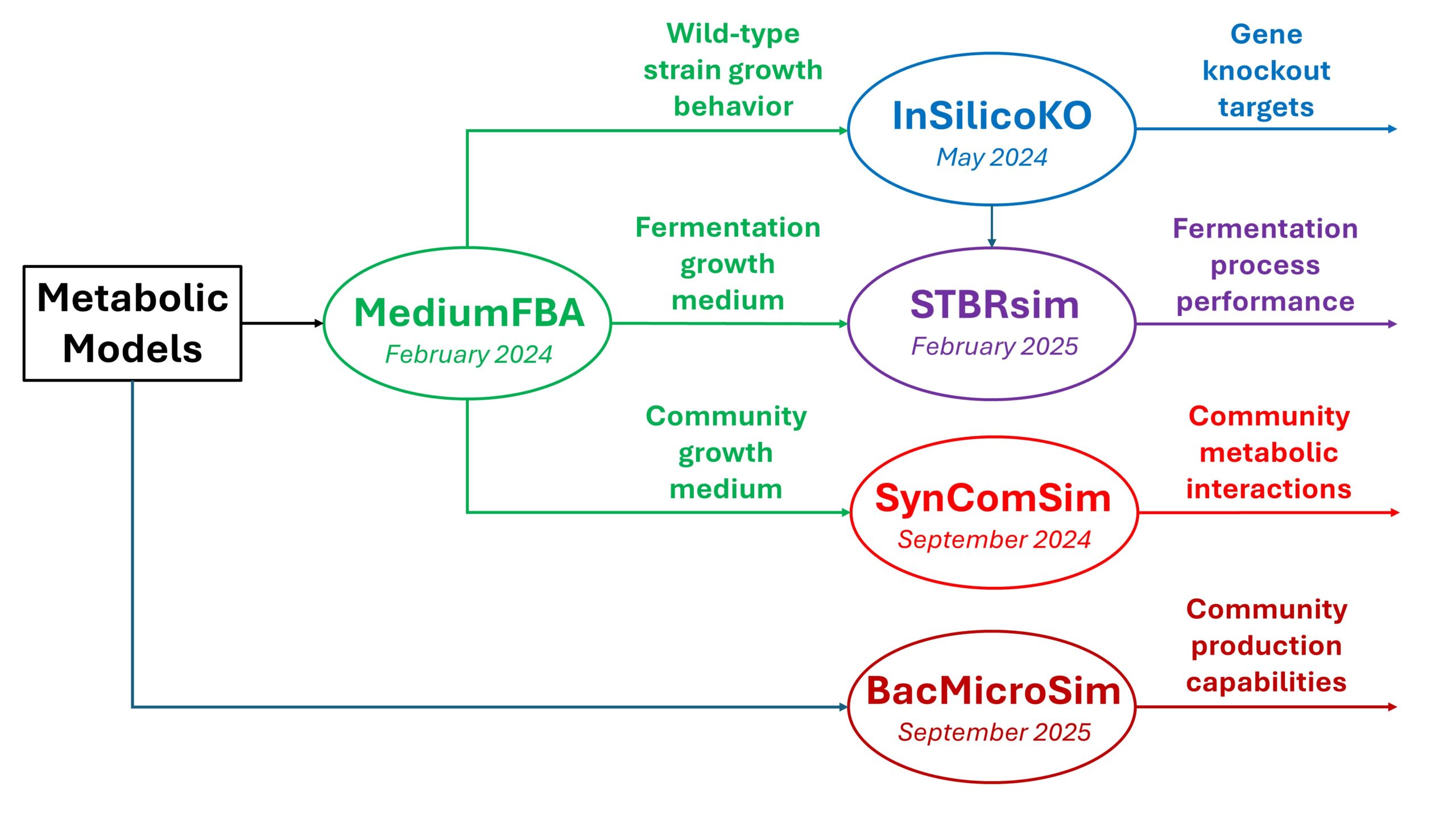
To address these challenges, In Silico Fermentation is developing a suite of free MATLAB GUI applications that enable academic and industrial researchers to quickly access the power of metabolic modeling and associated simulation methods. Starting with genome-scale metabolic reconstruction(s) of the microbial strain(s) of interest, these Apps allows the user to quickly define medium conditions, input strain-dependent metabolic parameters, simulate strain performance, interrogate strain interactions, and design improved mutant strains. The following five research Apps have been released.
- MediumFBA – Integrated Medium Design and Flux Balance Analysis for Genome-Scale Metabolic Models (first release February 2024).
- InSilicoKO – In Silico Design of Knockout Strains for Metabolite Overproduction using Genome-Scale Metabolic Models (second release May 2024).
- STBRsim – Dynamic Simulation of Stirred Tank Bioreactors Using Genome-Scale Metabolic Models (second release February 2025)
- SynComSim – Metabolic Simulation of Synthetic Microbial Communities using Genome-Scale Metabolic Models (first release September 2024)
- BacComSim – Simulation of Bacterial Communities Using Microbiome Read Data and Genome-Scale Metabolic Models (first release expected summer 2025)
These five Apps have been designed to allow seamless integration such that available metabolic models can be rapidly used for medium design, knockout strain design, fermentation process simulation, community composition prediction, metabolite production prediction, and community interaction interrogation.
Please contact me by email if you would like to learn more about these metabolic modeling Apps.