In Silico Fermentation can offer assistance in the development and testing of genome-scale metabolic reconstructions. In addition to serving as essential building blocks for our microbial bioreactor and community modeling tools, metabolic reconstructions represent a quantitative knowledge repository and valuable intellectual property. While my principal expertise is engineering applications of reconstruction-based models, I can assist in-house strain experts in developing metabolic reconstructions of your proprietary cell factories or targeted bacterial pathogens. Once curated with internal and publicly available data, the metabolic reconstruction can be interrogated individually or embedded within my other modeling tools.
Draft Reconstruction
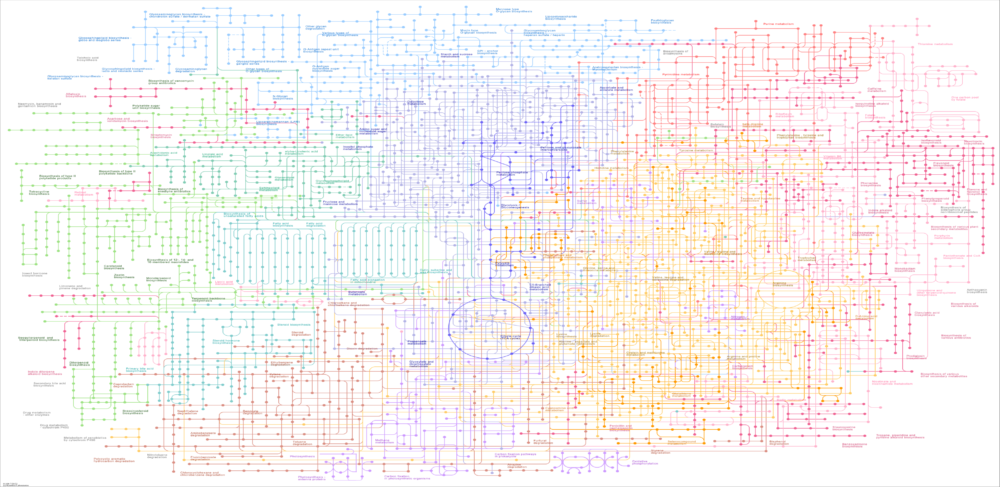
The starting point for any metabolic reconstruction project is the DNA sequence reads of the modeled strain. Using the ModelSEED computational pipeline within the DOE Systems Biology Knowledge Base (KBase), I can assemble and annotate the read data or simply upload an already annotated genome. A critical step in the reconstruction process is the computational definition of the growth media used for in vitro growth experiments. I can use KBase to translate the annotated genome and defined media into a preliminary metabolic pathway model, commonly known as a draft reconstruction. Close interactions with company strain experts are essential to evaluate the draft for incomplete, incorrect or missing pathways.
Curated Reconstruction
To develop a a high-fidelity reconstruction capable of accurate prediction, the draft model must undergo a manual curation process driven by internal strain experts. I can assist with the curation process by investigating reconstructions of phylogenetically related organisms and iteratively modifying and simulating the draft reconstruction through a series of build-test-learn cycles. Once the curation process is sufficiently advanced, I can work with fermentation engineers to design in vitro experiments for testing model predictions with respect to cellular growth rate, consumption rates of carbon sources and electron donors, and secretion rates of metabolic products. I can employ the curated reconstruction to perform a variety of computational experiments, including gene lethality tests and metabolic engineering studies , using the COBRA toolbox within MATLAB.
Please reach out to discuss your reconstruction development needs and my capabilities for metabolic model development!