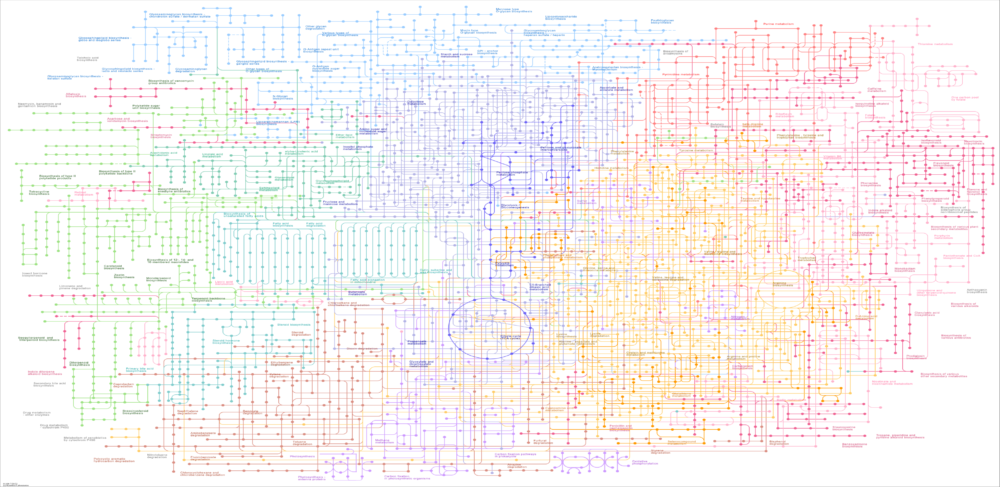
Genome-scale metabolic reconstructions have been leveraged to develop numerous computational methods for microbial system analysis and design. The incorporation of selected methods within software tools such as Constraint-Based Reconstruction and Analysis Toolbox (COBRA) allows experienced users to code specialized workflows to solve research-oriented problems. However, available tools are not well suited for many academic researchers and industrial practitioners who desire GUI-based software tools that are easily understood and used. Similarly, metabolic modeling is covered only in specialized biotechnology courses due to lack of pedagogical tools for introducing students to key modeling concepts in lieu of workflow coding exercises.
In Silico Fermentation is developing a suite of free MATLAB GUI applications that capture established metabolic modeling workflows for research and educational purposes. The research Apps allow academic researchers and industrial practitioners to quickly formulate, solve and analyze a broad range of metabolic modeling problems including flux balance analysis, growth medium design, microbial community simulation, gene knockout strain design, and bioreactor dynamic simulation. In parallel, we are developing educational Apps that provide instructors and students step-by-step guidance through the research App workflows within the context of specific applications such as bioreactor startup strategies.
Please contact me by email if you would like to learn more about these metabolic modeling Apps.