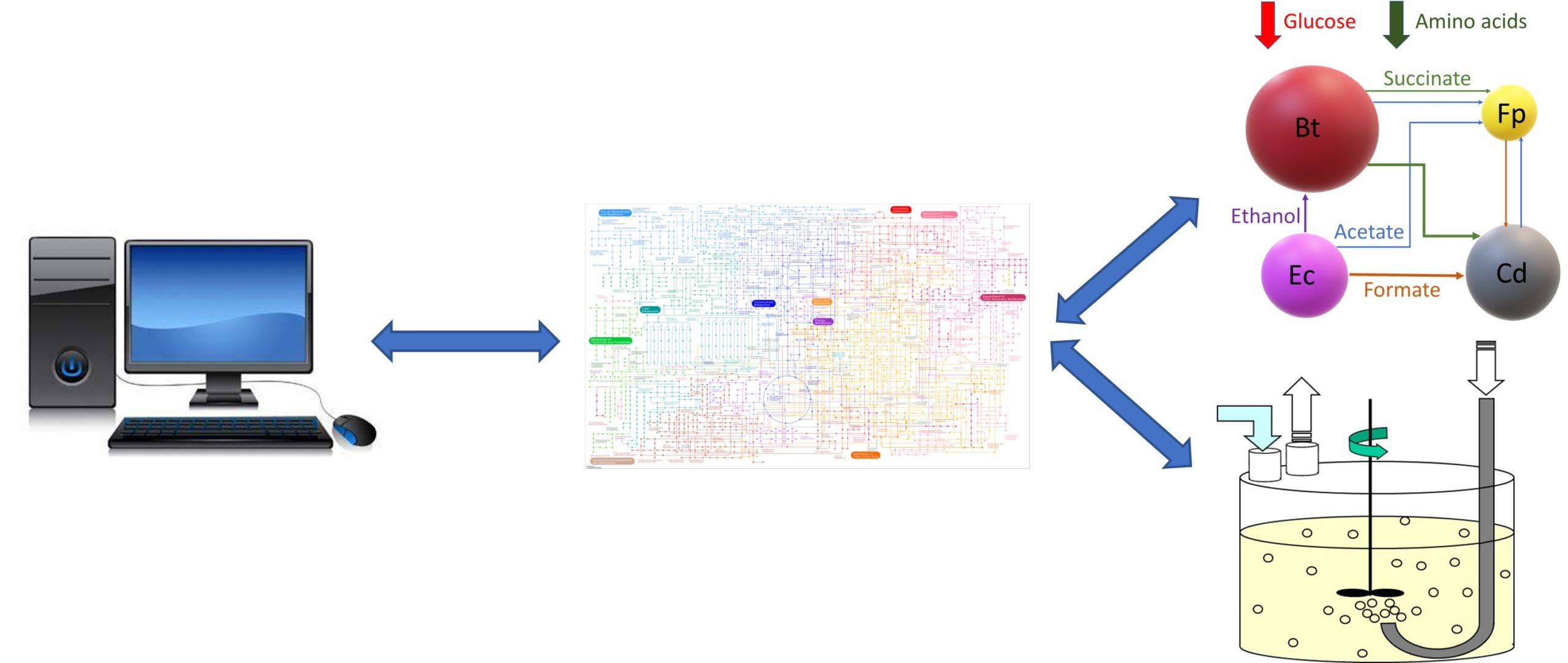
In Silico Fermentation offers vast experience in the metabolic modeling of microbial systems and fermentation processes. My novel computational workflow enables genome-scale metabolic reconstructions to be embedded within high-fidelity simulation models. Therefore, the same models used for detailed metabolic analysis of individual microbes can be integrated within larger scale system and process models to maximize modeling effort, value and impact.
Metabolic Reconstructions
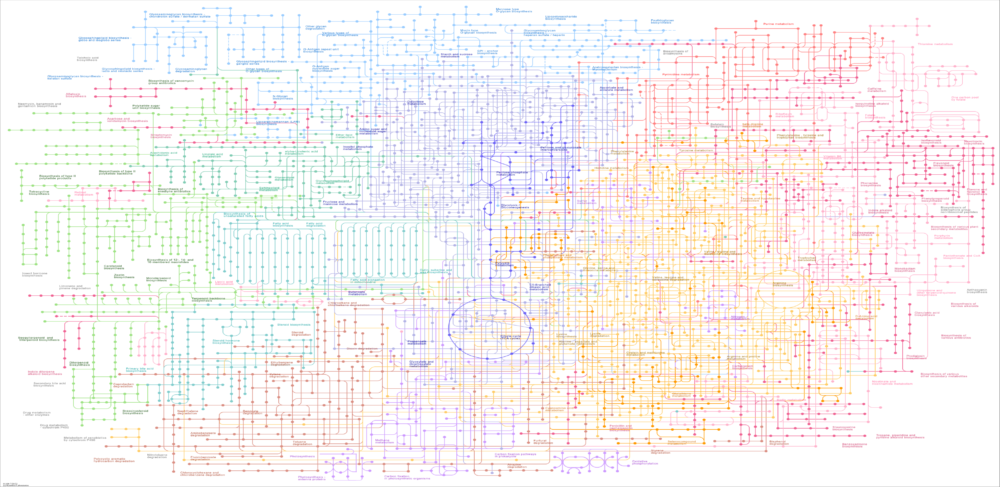
My modeling technology builds upon the increasing availability of metabolic reconstructions for diverse microbial strains. Some biotechnology companies have developed proprietary reconstructions for their particular microbial cell factories. Similarly, biomedical companies focused on infectious diseases may have in-house reconstructions for specific pathogenic strains. Moreover, standardized reconstruction pipelines and reliable computational tools are now available online to facilitate microbial metabolic modeling. I leverage these strain-specific reconstruction tools to develop predictive simulation models for diverse microbial applications.
Biochemical Reactors
My first general area of expertise is fermentation metabolic modeling. While metabolic reconstructions are commonly viewed only as a research tool, I have demonstrated the power of integrating microbial strain reconstructions within mass balance and transport equations to develop bioreactor simulation models. By doing so, inherently steady-state reconstruction are translated into dynamic fermenter models. This approach is extensible to a broad array of microbial bioreactor technologies including stirred tank reactors, bubble column fermenters, biofilm reactors and mixed-culture systems.
Microbial Communities
My second general area of expertise is microbial community metabolic modeling. The modeled communities may be naturally occurring where the participating taxa are quantified through metagenomics or synthetically engineered to have defined members. In either case, the microbiota model is developed by combining metabolic reconstructions of the participating taxa into a large-scale model following the flux balance analysis approach. The resulting models can be interrogated to understand metabolic interactions between community members and utilized for manipulation of community behavior. I can apply this approach to health- and disease-promoting bacterial communities associated with human hosts as well as to engineered microbial communities for sustainable fuel and chemical production.
Please take a few minutes to learn more about my expertise in metabolic reconstructions, bioreactor reactors and microbial communities!