STBRtutorial was released by In Silico Fermentation in January 2025. Please continue reading this page to learn more about the application and to download this free software.
In a typical introductory biochemical engineering course, students learn about unstructured modeling of microbial growth and basic differential equations models of stirred tank bioreactor (STBR) dynamics. While straightforward to describe in an undergraduate course, these simple models fail to capture the complex interplay between cellular metabolism and STBR operating conditions. As a result, classroom lectures often focus on methods for determination of growth-related parameters, such as Monod parameters and yield coefficients, rather than on STBR operation and dynamics. Moreover, this modeling approach quickly becomes intractable as the number of growth-limiting substrates, secreted products and operational stages (i.e., aerobic or anaerobic; batch, fed-batch or continuous) increases beyond the simplest case.
Genome-scale metabolic reconstructions can be exploited to develop more rigorous dynamic STBR models. However, concepts related to model development and simulation are well beyond the scope of an undergraduate course. As a result, the power of metabolic modeling methods are inaccessible to students who are tasked with learning basic microbial systems concepts rather than performing elaborate coding exercises.
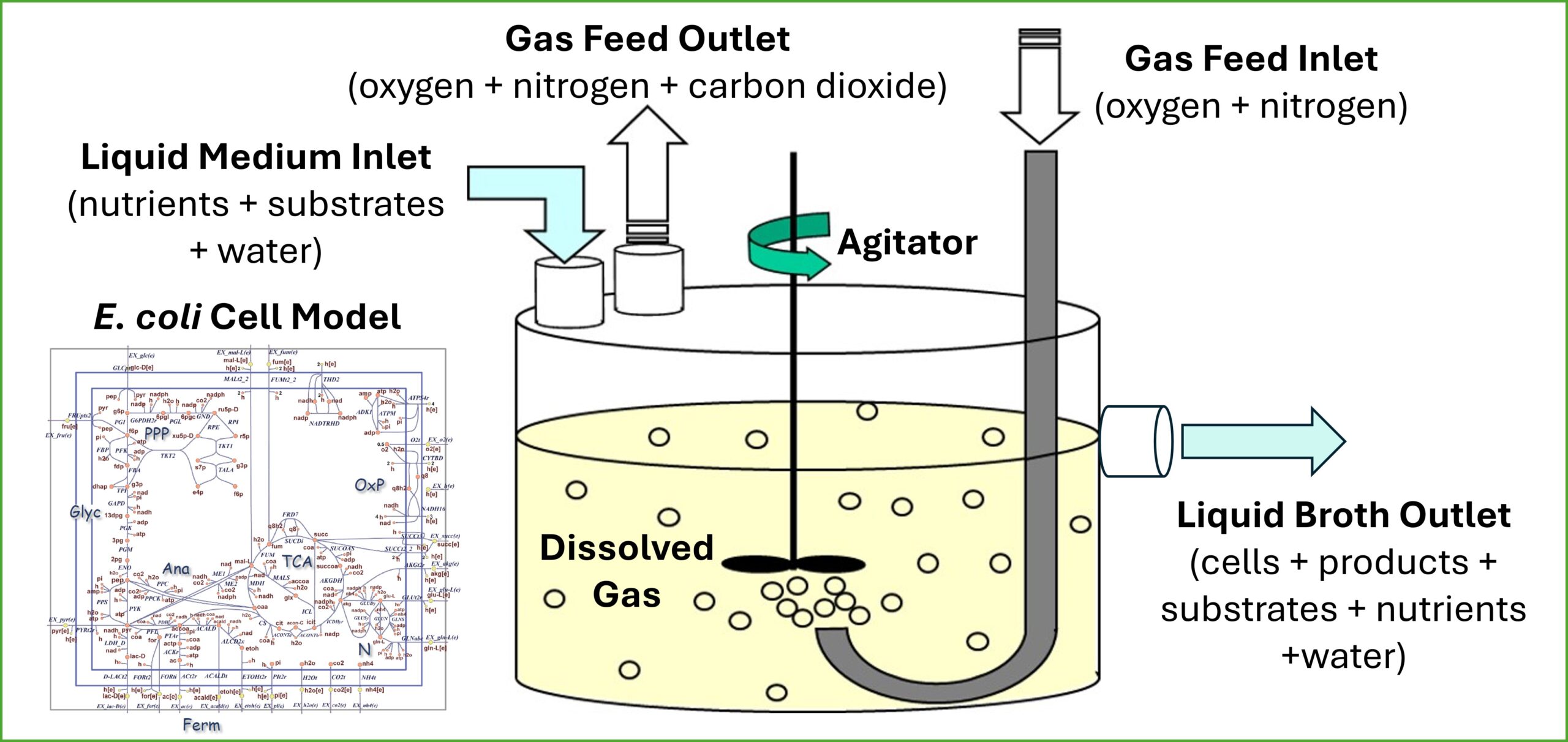
STBRtutorial is a MATLAB application that inconspicuously incorporates a core model of Escherichia coli metabolism within detailed tutorials that address important concepts about STBR operation and dynamics. Students are guided through an intuitive workflow in which they consider important aspects of defining a STBR problem, including the growth medium, growth-limiting substrates and secreted products, growth inhibition effects, bioreactor operating conditions including multiple stages for different liquid/gas feeding policies, gas-liquid mass transfer, and initial fermentation conditions. The GUI tool generates dynamic predictions of extracellular variables (i.e., biomass, substrate and product concentrations and liquid/gas phase volumes) and intracellular-extracellular transport fluxes (e.g., cellular growth rate, substrate uptake rates, product secretion rates).
STBRtutorial Version 1.0 provides two step-by-step tutorials that demonstrate how bioreactor operating policies affect STBR dynamic behavior. The first tutorial illustrates the impact of the liquid medium flow strategy on STBR startup dynamics under anaerobic conditions. Students compare different batch-to-continuous switching times on the approach to continuous flow steady state, demonstrating that startup dynamics are strongly impacted by the switching time and that the fastest dynamics are achieved when the batch-to-continuous switching occurs near glucose exhaustion.
The second tutorial introduces the concept of diauxic growth dynamics in an aerobic batch STBR. Students observe that E. coli can reassimilate secreted acetate following depletion of supplied glucose if sufficient dissolved oxygen is available for acetate oxidation. Different oxygen mass transfer coefficients are explored to demonstrate that diauxic growth dynamics are accelerated as oxygen mass transfer is increased. Conversely, students learn that diauxic growth does not occur for small oxygen mass transfer coefficients due to insufficient dissolved oxygen levels.
Each tutorial provides step-by-step instructions within the GUI to provide a totally self-contained training experience. The App state can be captured at any time during the tutorial so that students can reload their work at a later time to continue the example outside the tutorial. Near completion of the tutorial, students are instructed to use the included visualization App to compare and analyze the simulation runs performed. While a few guidelines are suggested at the end of each tutorial, the data analysis work is intended to be open ended so that students can explore the collected simulation data to understand how cellular metabolism and bioreactor operation interact to determine STBR dynamics. Plots generated in the visualization App can be saved for inclusion in submitted reports and oral presentations.
STBRtutorial has the following requirements:
- Version 2021a or newer version of MATLAB.
- Installed COBRA toolbox for MATLAB to provide access to the linear program solver glpk and other functions. Installation instructions are provided here.
STBRtutorial can be freely downloaded using the link below. The software is distributed under The GNU General Public License v3.0.

STBRtutorial is a MATLAB application that uses a core model of Escherichia coli metabolism to present detailed tutorials that address important concepts about stirred tank bioreactor (STBR) operation and dynamics.
The Zip file should be processed as follows:
- Download and unzip the file using Zip archive software.
- The unzipped file will contain the following files: (1) the main application, STBRtutoral_ver_1_0.mlappinstall; (2) the core E. coli metabolic model, e_coli_core.mat; (3) a jpeg graphics file loaded within the App, cstr_schematic_new; and (4) saved App states for the completed tutorials, startup_tutorial_case_final and diauxic_tutorial_case_final, to be used by the instructor as necessary.
- Save the files in the desired working directory.
- To install the STBRtutorial application, go to the Apps tab on the main Matlab page and click “Install App”. Select the saved STBRtutorial_ver_1_0.mlappinstall. The STBRtutorial application will appear as a new icon in the Apps tab.
- Launch the application by clicking on the icon.
Please contact me by email if you have any questions or feedback about STBRtutorial.