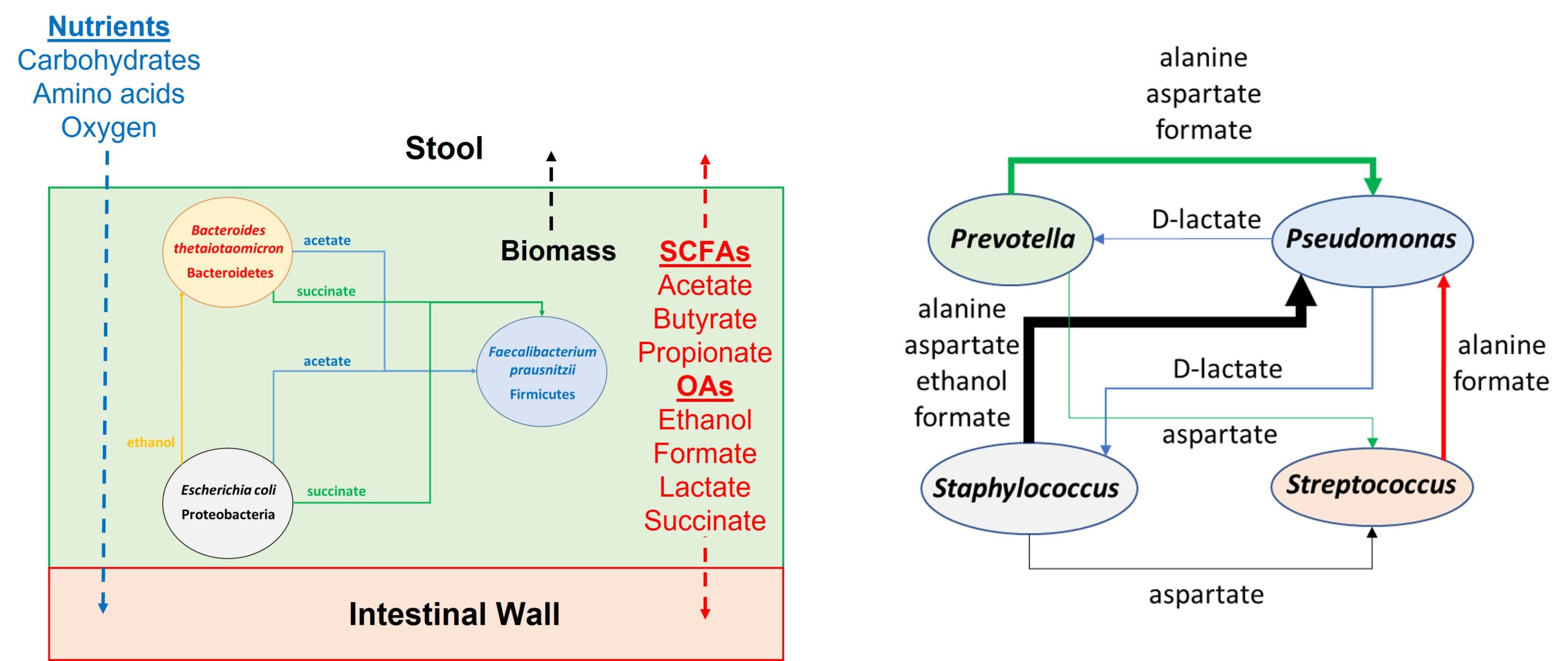
In Silico Fermentation provides expertise in microbiota metabolic modeling with applications to both natural and engineered microbial communities. These models provide a rigorous quantitative framework to predict the impact of engineered strains, invading pathogens and altered environments on community stability, structure and function. As described below, I have developed community models for a wide range of applications within the domains of renewable chemical production and human-host associated microbiomes. This experience can be translated to other in vitro and in vivo microbial systems studied by industrial practitioners and academic researchers.
Multispecies Biofilm Modeling
I have developed modeling tools to predict spatial organization and metabolic interactions of diverse microbes within multispecies biofilms. These models can be used to generate and test hypotheses regarding the roles of different species within health- or disease-promoting communities. Model interrogation allows the identification of metabolic targets within and between species to disrupt or promote biofilm formation. This expertise can be used to develop multispecies biofilm models for a wide variety of medical, environmental and synthetic systems. Some relevant publications include:
- Metabolic Modeling of a Chronic Wound Biofilm Consortium Predicts Spatial Partitioning of Bacterial Species
- Byproduct Cross Feeding and Community Stability in an In Silico Biofilm Model of the Gut Microbiome
- Metabolic Modeling of Clostridium difficile Associated Dysbiosis of the Gut Microbiota
Human Gut Microbiome Modeling
I have substantial experience developing community metabolic models for the human gut microbiota. Both small-scale biofilm and large-scale planktonic models can be constructed to understand the community conversion of food-derived nutrients to health- and disease-associated metabolites. Large-scale bacterial models can be generated from 16S rRNA gene sequence data to generate sample-specific communities. These combined capabilities allow the efficient development of community metabolic models for the gut microbiota and other human microbiomes. Some relevant publications include:
- Suboptimal Community Growth Mediated through Metabolite Crossfeeding Promotes Species Diversity in the Gut Microbiota
- Computational Modeling of the Gut Microbiota Reveals Putative Metabolic Mechanisms of Recurrent Clostridioides difficile Infection
- Microbiota Dysbiosis in Inflammatory Bowel Diseases: In silico Investigation of the Oxygen Hypothesis
Disease-Associated Microbiome Modeling
I offer expertise in the the development of community metabolic models to perform in silico analyses of disease-associated microbiota. The availability of 16S rRNA gene sequence data allows the development of sample-specific models and computational interrogation of healthy- versus disease-associated communities. These microbiome modeling tools been successfully applied to bacterial communities involved in human infections present in the gut, chronic wounds and the cystic fibrosis airway. This experience can be leveraged to analyze bacterial communities involved in a wide variety of human infections and disease processes. Some relevant publications include:
- Metabolic Modeling of Cystic Fibrosis Airway Communities Predicts Mechanisms of Pathogen Dominance
- Metabolic Modeling of Chronic Wound Microbiota Predicts Mutualistic Interactions that Drive Community Composition
- Interrogation of the Perturbed Gut Microbiota in Gouty Arthritis Patients Through In silico Metabolic Modeling
Please contact me to discuss your microbiota modeling needs and learn more about my computational capabilities!