In Silico Fermentation offers unique capabilities for microbial bioreactor modeling based on genome-scale metabolic reconstructions. The development of rigorous dynamic models that integrate detailed metabolic descriptions with process-relevant transport mechanisms provides a powerful computational platform for process innovation, improvement and optimization. As described below, my modeling tools have been applied to a broad array of bioreactor technologies including stirred tanks, bubble columns, biofilms and mixed cultures. I can readily translate this experience to other bioreactor modeling problems encountered in industrial manufacturing, pilot development and initial conceptualization.
Gas Fermentation Modeling
I pioneered the use of metabolic reconstructions for gas fermentation simulation. My models are equally applicable to stirred tank fermenters used for laboratory analyses and bubble column reactors developed for large-scale production. Validated bioreactor models can be leveraged to improve process performance through in silico metabolic engineering studies, simulation of multiphase reactor dynamics, and optimization of reactor operating conditions. These bioreactor modeling tools are readily extensible to a broad range of gas fermentation processes. Some relevant publications include:
- Metabolic Modeling of Synthesis Gas Fermentation in Bubble Column Reactors
- In silico Metabolic Engineering of Clostridium ljungdahlii for Synthesis Gas Fermentation
- Experimental Testing of a Spatiotemporal Metabolic Model for Carbon Monoxide Fermentation with Clostridium autoethanogenum
Mixed-Culture Bioreactor Modeling
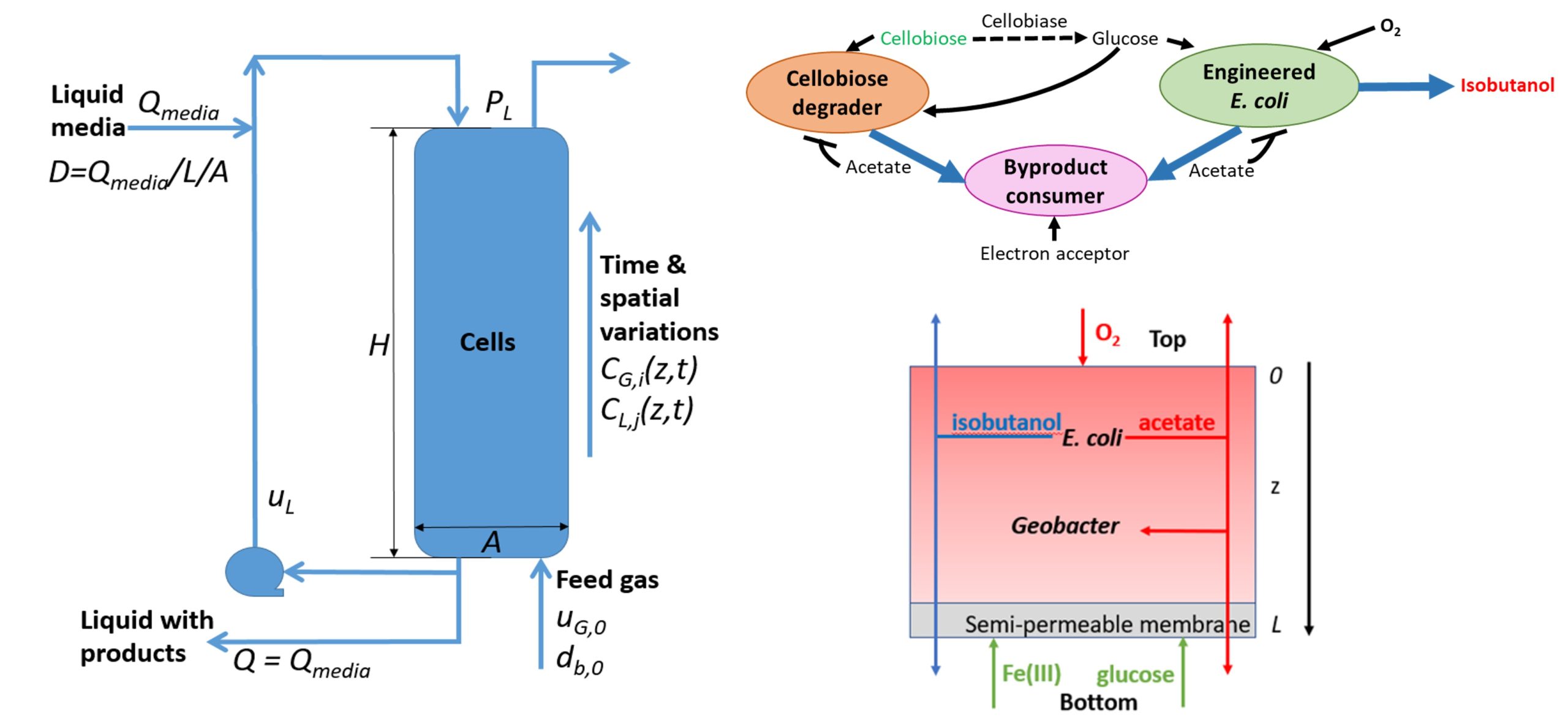
I offer extensive experience developing mixed-culture metabolic models for bioconversion applications. My bioreactor modeling tools can be applied to defined microbial communities to decipher and direct species interactions. Dynamic process models can be developed for stirred tank, bubble column and biofilm reactors to predict and optimize fermentation performance. These modeling methods are applicable to a wide range of bioconversion processes for sustainable production of fuels and chemicals. Some relevant publications include:
- Dynamic Metabolic Modeling of a Microaerobic Yeast Co-culture: Predicting and Optimizing Ethanol Production from Glucose/Xylose Mixture
- Dynamic Model-Based Analysis of Furfural and HMF Detoxification by Pure and Mixed Batch Cultures of S. cerevisiae and S. stipitis
- Metabolic Modeling of Bacterial Co-culture Systems Predicts Enhanced Carbon Monoxide-to-Butyrate Conversion Compared to Monoculture Systems
Engineered Biofilm Modeling
I pioneered the development of biofilm metabolic models for renewable chemical synthesis. These models can be used for spatiotemporal simulation of diffusion-limited biofilm metabolism and to computationally identify promising combinations of microbial species for a particular bioconversion task. The modeling tools are extensible to a broad array of microbial species, renewable feedstocks and target products. Some relevant publications include:
- Spatiotemporal Modeling of Microbial Metabolism
- In silico Metabolic Design of Two-species Biofilm Systems Predicts Enhanced Biomass Production and Biochemical Synthesis
- In silico Analysis of Synthetic Multispecies Biofilms for Cellobiose-to-Isobutanol Conversion Reveals Design Principles for Stable and Productive Communities
Please reach out to discuss your fermentation modeling needs and my powerful computational tools!