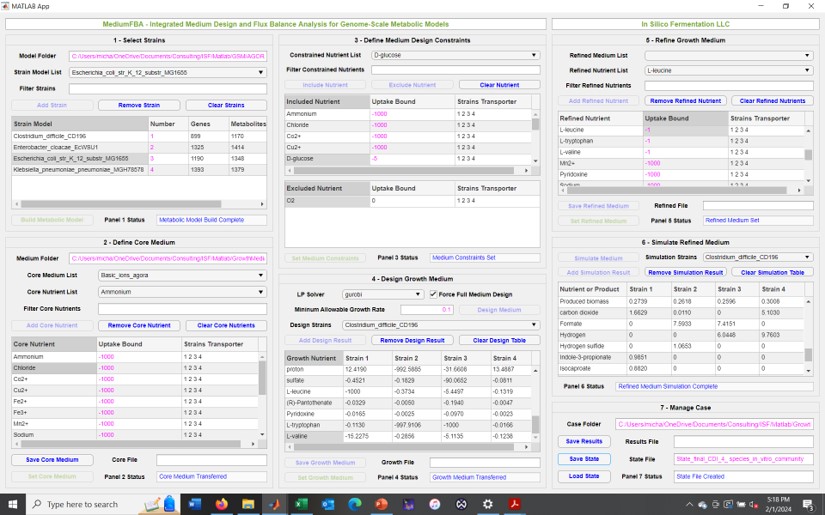
In Silico Fermentation offers unique capabilities for developing MATLAB applications to standardize and simplify computational modeling workflows. I have particular expertise in App development for metabolic modeling workflows involving genome-scale metabolic reconstructions and linear programming computations. In 2024, I will release free MATLAB Apps for growth medium design, in silico metabolic engineering, fermentation process simulation and microbial community simulation to advance industrial utilization of metabolic modeling. Despite the generality of these four Apps, my experience suggests that many biotechnology and biomedical companies have more specialized workflows that can benefit from strategic App development. In Silico Fermentation can work with industrial partners to conceptualize, design, construct, test and commission Apps for a wide range of workflows associated with data analysis, computational modeling, systems optimization and process control.
Microbial Metabolic Modeling
In Silico Fermentation offers established expertise in developing MATLAB apps for implementation of metabolic modeling workflows. Starting with genome-scale metabolic reconstructions of the modeled strain(s), I can build flexible Apps that perform medium design, flux balance analysis, gene knockout mutant design, and synthetic and host-associated community simulations. More generally, Apps can be developed for a wide range of metabolic model types ranging from simple unstructured growth models to the most sophisticated whole-cell models. If your metabolic modeling workflows needs to be implemented in a robust and reproducible manner, I can provide the expertise lacking in your organization.
Fermentation Process Simulation
In Silico Fermentation offers substantial experience in dynamic modeling and simulation of microbial fermentation processes with particular expertise in submerged liquid and bubble column bioreactors. When combined with my expertise in MATLAB App development, I develop a broad range of fermentation process simulation Apps that either standardize existing internal workflows or establish new modeling workflows. My freely distributed App STBRsim demonstrates how a genome-scale metabolic reconstruction can be used as a rigorous scaffold for developing detailed stirred tank bioreactor simulations. Similarly, I can develop fermentation simulation Apps based on other model types to accelerate the innovation cycle from strain engineering to production scaleup.
Biosystems Optimization and Control
While In Silico Fermentation specializes in microbial metabolic modeling, I can develop MATLAB Apps to encode workflows in related areas of bioprocess systems engineering. I have substantial experience with model parameter estimation from experimental data, model-based optimization of reactor operating conditions, and reactor feedback control ranging from simple PID methods to more sophisticated model-based approaches. I have the ability to translate computational workflows within these domains to intuitive and robust Apps that eliminate the need for specific in-house expertise. Whatever your bioprocess engineering needs, I likely can developed the software tools required to advance your efforts.